|
Tree sort algorithm for the prediction of siRNA with minimized off-target hybridization
Jonathan Bode, Maria Emelianenko, Ganiraju Manyam, Ancha Baranova
This is a collaborative project between
School of Systems Biology, College of Science, George Mason University, Fairfax, VA
Department of Mathematics, College of Science, George Mason University, Fairfax, VA
This manuscript is published in BMC Research Notes in 2011 "An efficient algorithm for systematic analysis of nucleotide strings suitable for siRNA design".
The "off-target" silencing effect hinders the development of siRNA-based therapeutic and research applications. Existing solutions for finding possible locations of siRNA seats within a large database of genes are either too slow, miss a portion of the targets, or are simply not designed to handle a very large number of queries. We propose a new approach that reduces the computational time as compared to existing techniques.
FINDINGS:
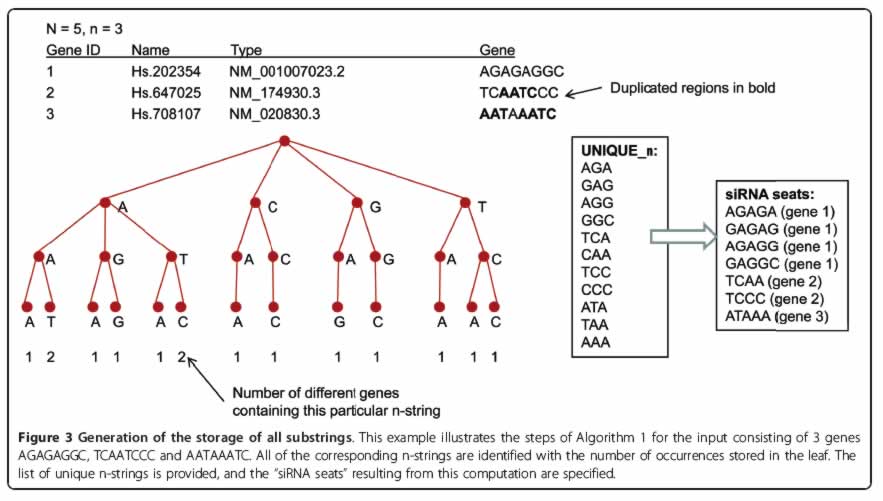
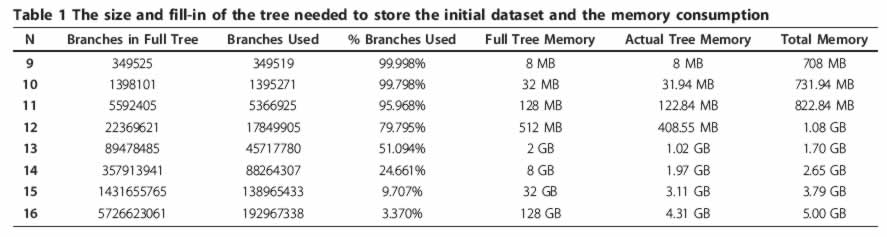
The proposed method employs tree-based storage in a form of a modified truncated suffix tree to sort all possible short string substrings within given set of strings (i.e. transcriptome). Using the new algorithm, we pre-computed a list of the best siRNA locations within each human gene ("siRNA seats"). siRNAs designed to reside within siRNA seats are less likely to hybridize off-target. These siRNA seats could be used as an input for the traditional "set-of-rules" type of siRNA designing software. The list of siRNA seats is available through a publicly available database located at http://web.cos.gmu.edu/~gmanyam/siRNA_db/search.php
CONCLUSIONS:
In attempt to perform top-down prediction of the human siRNA with minimized off-target hybridization, we developed an efficient algorithm that employs suffix tree based storage of the substrings. Applications of this approach are not limited to optimal siRNA design, but can also be useful for other tasks involving selection of the characteristic strings specific to individual genes. These strings could then be used as siRNA seats, as specific probes for gene expression studies by oligonucleotide-based microarrays, for the design of molecular beacon probes for Real-Time PCR and, generally, any type of PCR primers.
Please note that this project represent further extension of the earlier project described at:
Small Interference (si) RNAs with High Specificity to the Target: a Systematic Approach to Design by CRM Algorithm
and published in Molecular Biology (Moscow) 2008 Jan-Feb;42(1):163-71.
English translation of this paper is here:
|